sklearn.metrics.silhouette_score¶
- sklearn.metrics.silhouette_score(X, labels, metric='euclidean', sample_size=None, random_state=None, **kwds)[source]¶
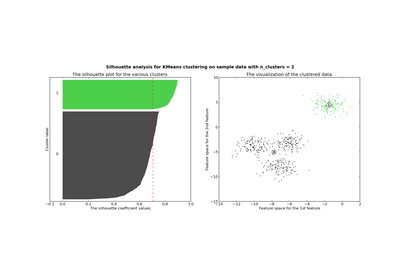
Compute the mean Silhouette Coefficient of all samples.
The Silhouette Coefficient is calculated using the mean intra-cluster distance (a) and the mean nearest-cluster distance (b) for each sample. The Silhouette Coefficient for a sample is (b - a) / max(a, b). To clarify, b is the distance between a sample and the nearest cluster that the sample is not a part of. Note that Silhouette Coefficent is only defined if number of labels is 2 <= n_labels <= n_samples - 1.
This function returns the mean Silhouette Coefficient over all samples. To obtain the values for each sample, use silhouette_samples.
The best value is 1 and the worst value is -1. Values near 0 indicate overlapping clusters. Negative values generally indicate that a sample has been assigned to the wrong cluster, as a different cluster is more similar.
Read more in the User Guide.
Parameters: X : array [n_samples_a, n_samples_a] if metric == “precomputed”, or, [n_samples_a, n_features] otherwise
Array of pairwise distances between samples, or a feature array.
labels : array, shape = [n_samples]
Predicted labels for each sample.
metric : string, or callable
The metric to use when calculating distance between instances in a feature array. If metric is a string, it must be one of the options allowed by metrics.pairwise.pairwise_distances. If X is the distance array itself, use metric="precomputed".
sample_size : int or None
The size of the sample to use when computing the Silhouette Coefficient on a random subset of the data. If sample_size is None, no sampling is used.
random_state : integer or numpy.RandomState, optional
The generator used to randomly select a subset of samples if sample_size is not None. If an integer is given, it fixes the seed. Defaults to the global numpy random number generator.
`**kwds` : optional keyword parameters
Any further parameters are passed directly to the distance function. If using a scipy.spatial.distance metric, the parameters are still metric dependent. See the scipy docs for usage examples.
Returns: silhouette : float
Mean Silhouette Coefficient for all samples.
References
[R191] Peter J. Rousseeuw (1987). “Silhouettes: a Graphical Aid to the Interpretation and Validation of Cluster Analysis”. Computational and Applied Mathematics 20: 53-65. [R192] Wikipedia entry on the Silhouette Coefficient