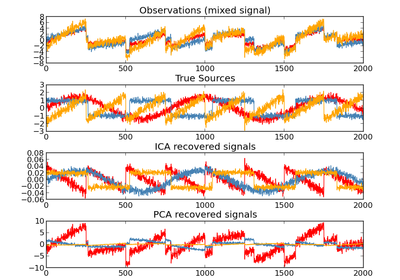
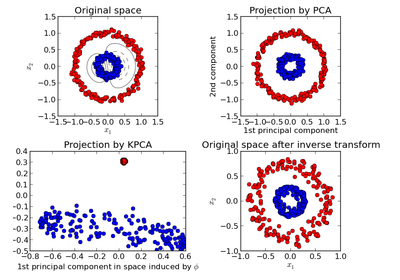
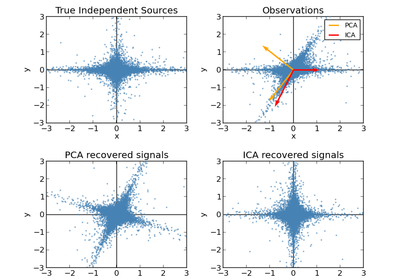
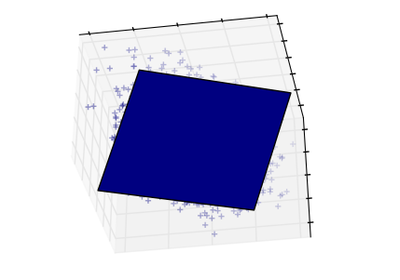
sklearn.decomposition.PCA¶
- class sklearn.decomposition.PCA(n_components=None, copy=True, whiten=False)[source]¶
Principal component analysis (PCA)
Linear dimensionality reduction using Singular Value Decomposition of the data and keeping only the most significant singular vectors to project the data to a lower dimensional space.
This implementation uses the scipy.linalg implementation of the singular value decomposition. It only works for dense arrays and is not scalable to large dimensional data.
The time complexity of this implementation is O(n ** 3) assuming n ~ n_samples ~ n_features.
Read more in the User Guide.
Parameters: n_components : int, None or string
Number of components to keep. if n_components is not set all components are kept:
n_components == min(n_samples, n_features)
if n_components == ‘mle’, Minka’s MLE is used to guess the dimension if 0 < n_components < 1, select the number of components such that the amount of variance that needs to be explained is greater than the percentage specified by n_components
copy : bool
If False, data passed to fit are overwritten and running fit(X).transform(X) will not yield the expected results, use fit_transform(X) instead.
whiten : bool, optional
When True (False by default) the components_ vectors are divided by n_samples times singular values to ensure uncorrelated outputs with unit component-wise variances.
Whitening will remove some information from the transformed signal (the relative variance scales of the components) but can sometime improve the predictive accuracy of the downstream estimators by making there data respect some hard-wired assumptions.
Attributes: components_ : array, [n_components, n_features]
Principal axes in feature space, representing the directions of maximum variance in the data. The components are sorted by explained_variance_.
explained_variance_ : array, [n_components]
The amount of variance explained by each of the selected components.
explained_variance_ratio_ : array, [n_components]
Percentage of variance explained by each of the selected components.
If n_components is not set then all components are stored and the sum of explained variances is equal to 1.0.
mean_ : array, [n_features]
Per-feature empirical mean, estimated from the training set.
Equal to X.mean(axis=1).
n_components_ : int
The estimated number of components. Relevant when n_components is set to ‘mle’ or a number between 0 and 1 to select using explained variance.
noise_variance_ : float
The estimated noise covariance following the Probabilistic PCA model from Tipping and Bishop 1999. See “Pattern Recognition and Machine Learning” by C. Bishop, 12.2.1 p. 574 or http://www.miketipping.com/papers/met-mppca.pdf. It is required to computed the estimated data covariance and score samples.
See also
Notes
For n_components=’mle’, this class uses the method of Thomas P. Minka: Automatic Choice of Dimensionality for PCA. NIPS 2000: 598-604
Implements the probabilistic PCA model from: M. Tipping and C. Bishop, Probabilistic Principal Component Analysis, Journal of the Royal Statistical Society, Series B, 61, Part 3, pp. 611-622 via the score and score_samples methods. See http://www.miketipping.com/papers/met-mppca.pdf
Due to implementation subtleties of the Singular Value Decomposition (SVD), which is used in this implementation, running fit twice on the same matrix can lead to principal components with signs flipped (change in direction). For this reason, it is important to always use the same estimator object to transform data in a consistent fashion.
Examples
>>> import numpy as np >>> from sklearn.decomposition import PCA >>> X = np.array([[-1, -1], [-2, -1], [-3, -2], [1, 1], [2, 1], [3, 2]]) >>> pca = PCA(n_components=2) >>> pca.fit(X) PCA(copy=True, n_components=2, whiten=False) >>> print(pca.explained_variance_) [ 6.6162... 0.05038...] >>> print(pca.explained_variance_ratio_) [ 0.99244... 0.00755...]
Methods
fit(X[, y]) Fit the model with X. fit_transform(X[, y]) Fit the model with X and apply the dimensionality reduction on X. get_covariance() Compute data covariance with the generative model. get_params([deep]) Get parameters for this estimator. get_precision() Compute data precision matrix with the generative model. inverse_transform(X) Transform data back to its original space using n_components_. score(X[, y]) Return the average log-likelihood of all samples. score_samples(X) Return the log-likelihood of each sample. set_params(**params) Set the parameters of this estimator. transform(X) Apply the dimensionality reduction on X. - fit(X, y=None)[source]¶
Fit the model with X.
Parameters: X: array-like, shape (n_samples, n_features) :
Training data, where n_samples in the number of samples and n_features is the number of features.
Returns: self : object
Returns the instance itself.
- fit_transform(X, y=None)[source]¶
Fit the model with X and apply the dimensionality reduction on X.
Parameters: X : array-like, shape (n_samples, n_features)
Training data, where n_samples is the number of samples and n_features is the number of features.
Returns: X_new : array-like, shape (n_samples, n_components)
- get_covariance()[source]¶
Compute data covariance with the generative model.
cov = components_.T * S**2 * components_ + sigma2 * eye(n_features) where S**2 contains the explained variances.
Returns: cov : array, shape=(n_features, n_features)
Estimated covariance of data.
- get_params(deep=True)[source]¶
Get parameters for this estimator.
Parameters: deep: boolean, optional :
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: params : mapping of string to any
Parameter names mapped to their values.
- get_precision()[source]¶
Compute data precision matrix with the generative model.
Equals the inverse of the covariance but computed with the matrix inversion lemma for efficiency.
Returns: precision : array, shape=(n_features, n_features)
Estimated precision of data.
- inverse_transform(X)[source]¶
Transform data back to its original space using n_components_.
Returns an input X_original whose transform would be X.
Parameters: X : array-like, shape (n_samples, n_components)
New data, where n_samples is the number of samples and n_components is the number of components. X represents data from the projection on to the principal components.
Returns: X_original array-like, shape (n_samples, n_features) :
- score(X, y=None)[source]¶
Return the average log-likelihood of all samples.
See. “Pattern Recognition and Machine Learning” by C. Bishop, 12.2.1 p. 574 or http://www.miketipping.com/papers/met-mppca.pdf
Parameters: X: array, shape(n_samples, n_features) :
The data.
Returns: ll: float :
Average log-likelihood of the samples under the current model
- score_samples(X)[source]¶
Return the log-likelihood of each sample.
See. “Pattern Recognition and Machine Learning” by C. Bishop, 12.2.1 p. 574 or http://www.miketipping.com/papers/met-mppca.pdf
Parameters: X: array, shape(n_samples, n_features) :
The data.
Returns: ll: array, shape (n_samples,) :
Log-likelihood of each sample under the current model
- set_params(**params)[source]¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The former have parameters of the form <component>__<parameter> so that it’s possible to update each component of a nested object.
Returns: self :
- transform(X)[source]¶
Apply the dimensionality reduction on X.
X is projected on the first principal components previous extracted from a training set.
Parameters: X : array-like, shape (n_samples, n_features)
New data, where n_samples is the number of samples and n_features is the number of features.
Returns: X_new : array-like, shape (n_samples, n_components)